Allows user to combine supplied structures and calculated scattering profiles into a single set of new files.
The Merge Utilities module is accessible from the Tools section of the main menu.
The purpose of the module is combine the results from multiple runs to consilidate files to enable analysis as if they were derived from a single run.
The default usage will merge all structures / SAS profiles. There are additional options to merge based on a user supplied weight_files (one per run you wish to merge) or to merge based on a user supplied sampling frequency.
An example of each use case is shown below.
Typical usage is to combine structures from DCD files and corresponding SAS profiles.
This module makes new copies of the data into a single DCD file and/or a new directory containging SAS profiles.
The default output file format is DCD. If you wish to save the merged coordinates as a PDB file then type in a filename with .pdb at the end. For large numbers of frames we recommend saving your data in the DCD format (~seven fold smaller file size).
Merging runs preserves the association so that each DCD frame corresponds to it's original SAS profile. Numbering in the new files is based on 1 and is continuous and it is not related to the original numbering.
Structures / SAS profiles are counted starting at 1. Some processing and visualization programs start counting at 0.
The "weights" file format is that generated (Chi-Square Filter) and used (Density Plot) in this and other modules.
The "weights" file format is used in this module to merge specific strucutres that fit the cutoff used to generate these files in (Chi-Square Filter). For example, one can use the "weights" file to merge subsets of structures / SAS profiles with the reduced chi-square values less than a certain value.
To merge structures and/or SAS profiles one selects the appropriate checkbox(es).
If you are merging structures and SAS profiles they should be a matched set for each pair of runs you are merging. In other words, the SAS profiles were calcualted from the structures in your input trajectory file. The program will not check if this is true or not. Different runs that are being merged can have different number of structures / SAS profiles from other runs, but must have the same number of structures / SAS profiles for a given run.
SAS profiles are selected by their location on the server. They must exist prior to running this module as there is no utility to upload a directory of SAS profiles to the server.
SAS profiles generated from SasCalc, Xtal2Sas, Cryson, or Crysol are currently supported. NOTE that retired SAS calculators may not be supported in the future.
One cannot merge SAS profiles created using different SAS calculators.
SAS profiles calculated using SasCalc can be merged simulaneously for multiple contrasts by selecting the top level sascalc directories. See Case 4 below for details.
This example merges structures and SAS profiles using each of the use cases mentioned above, assuming that the SAS profiles were calculated using SasCalc. If a different SAS module was used to calculate the SAS profiles, the SAS type and SAS data path must be selected accordingly. Usage where only structures or only SAS profiles are merged are not shown, but the input fields for the individual cases does not change in these conditions.
Note, that this example requires you to generate two sets of SAS profiles independently using one of the supported SAS modules in Calculate.
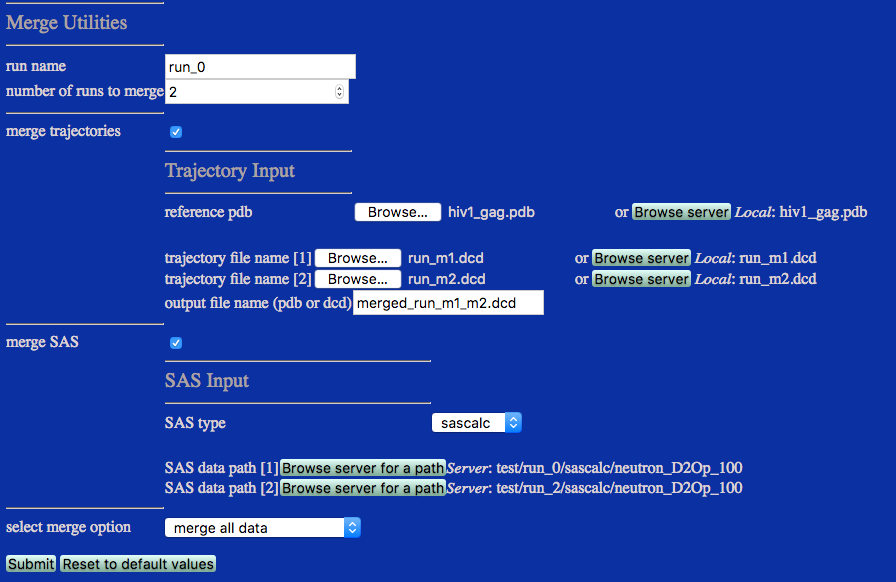
This example merges all structures from trajectory files uploaded by the user and copies SAS profiles from pre-existing folders on the server. Alternatively, trajectory files could be accessed directly from ther server.
Note that the sascalc/neutron_D2Op_100 directories are selected to merge SAS profiles only for that particular contrast.

run name: user defined name of folder that will contain the results.
extract trajectory selecting this checkbox enables the Trajectory Input fields
trajectory filename: DCD or PDB file with coordinates that will be merged. One input field for each run to be merged will be displayed.
output file name (pdb or dcd): Name of output PDB or DCD file with the merged coordinates.
extract SAS selecting this checkbox enables the SAS Input fields
SAS type select the name of the SAS calculator that was used to generate the SAS profiles that you are going to merge.
SAS data path select the directories that contain SAS profiles. One input field for each run to be merged will be displayed.
select merge option Pull down menu with options for 1. merge all data, 2. merge using weight files, 3. merge periodic. The option choice will spawn appropriate input field.
This example extracts frames and SAS profiles using information from weight files from each run. In the box below only the first few lines are shown for one of the two weight files.
# file generated on Sun Apr 12 18:05:32 2015
# structure, X2, weight
1 10.394808 0.000000
2 8.041527 0.000000
3 8.792317 0.000000
4 8.217490 0.000000
5 6.896897 1.000000
6 7.555859 1.000000
7 8.220753 0.000000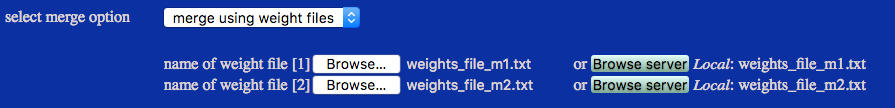
This example extracts frames and SAS profiles using the input sampling frequency.

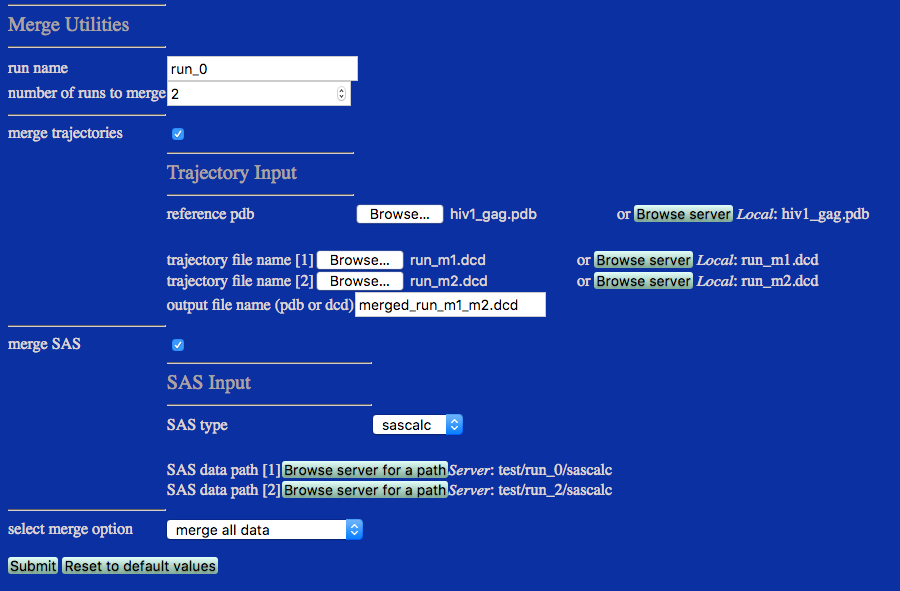
This example merges all structures from trajectory files uploaded by the user and copies SAS profiles from multiple pre-existing folders on the server that were created from SasCalc calculations at different contrasts. Alternatively, trajectory files could be accessed directly from ther server.
Note the difference in SAS data paths from Case 1 above. The top level /sascalc directories are selected rather than the contrast-specific directories, /sascalc/neutron_D2Op_100.
Results will be written to a new directory within the given "run name". For example, in the figure it is noted that the structures were saved files within the current project directory within the chosen "run name" directory. SAS profiles are written to a directory named by the SAS calculator used to create the original files.
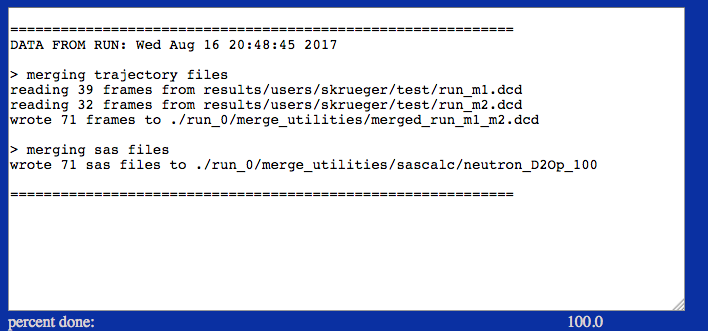
./run_0/merge_utilities/merged_run_m1_m2.dcd
./run_0/merge_utilities/sascalc/neutron_D2Op_100
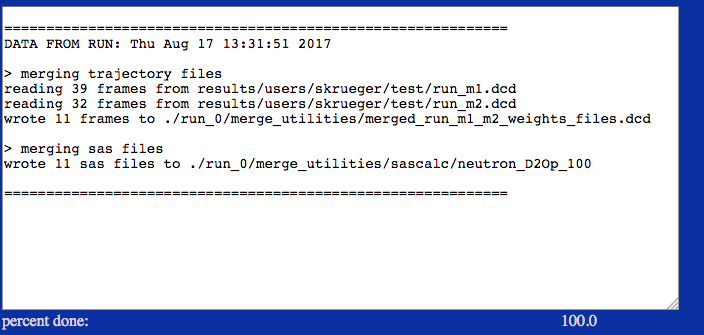
./run_0/merge_utilities/merged_run_m1_m2_weights_file.dcd
./run_0/merge_utilities/sascalc/neutron_D2Op_100
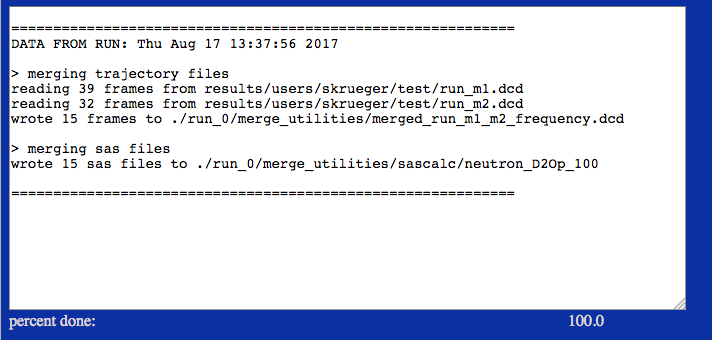
./run_0/merge_utilities/merged_run_m1_m2_frequency.dcd
./run_0/merge_utilities/sascalc/neutron_D2Op_100
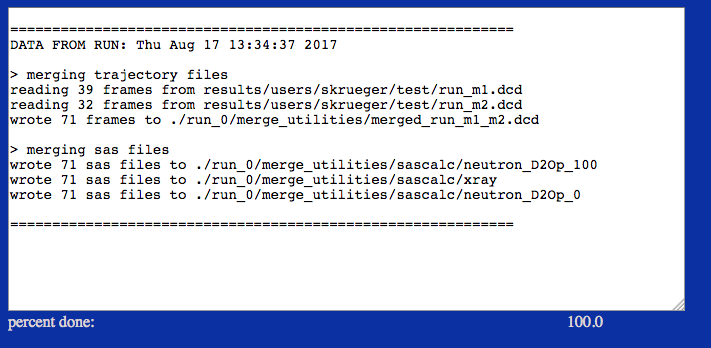
Note the difference between this output and that for Case 1 above. The SAS files are merged from all sets of directories below the top level /sascalc directory, i.e., /neutron_D2Op_100, /neutron_D2Op_0 and /xray.
./run_0/merge_utilities/merged_run_m1_m2.dcd
./run_0/merge_utilities/sascalc/neutron_D2Op_100
./run_0/merge_utilities/sascalc/xray
./run_0/merge_utilities/sascalc/neutron_D2Op_0
None
input files
hiv1_gag.pdb
run_m1.dcd
run_m2.dcd
weights_file_m1.txt
weights_file_m2.txt
output files
merged_run_m1_m2.dcd
merged_run_m1_m2_weights_file.dcd
merged_run_m1_m2_frequency.dcd
NOTE: input and output SAS profiles are not available for download since they cannot be uploaded to test the module.
Only PDB and DCD file formats are supported. SAS profiles must exist on the server as there is no option to upload a folder containg such files.
Not published.